Publications
People
Understanding the environmental persistence of endemic and emerging pathogens and identifying mechanisms of inactivation
Viral infections exhibit distinct seasonality. For instance, influenza virus largely displays winter seasonality in temperate regions, while parainfluenza virus typically causes waves of infection in the summer and fall. While factors such as virus persistence and transmissibility and changes in host response mechanisms under different environmental conditions are hypothesized causes of seasonality, the dynamics governing virus spread remain unknown.
Our lab is working to comprehensively assess and compare the persistence of important but distinct enveloped and nonenveloped viral pathogens in droplets and aerosols under a range of environmental conditions. This information will improve our understanding of the mechanisms driving environmental inactivation of particular microbes of interest. Using experimental data on a range of viruses, we can develop computational models to predict how respiratory virus persistence is impacted by environmental and viral (e.g., structural) factors. This work will define engineering treatments targeted at specific pathogens to rapidly reduce infectious viral loads in air or on surfaces during intervals of elevated community spread.
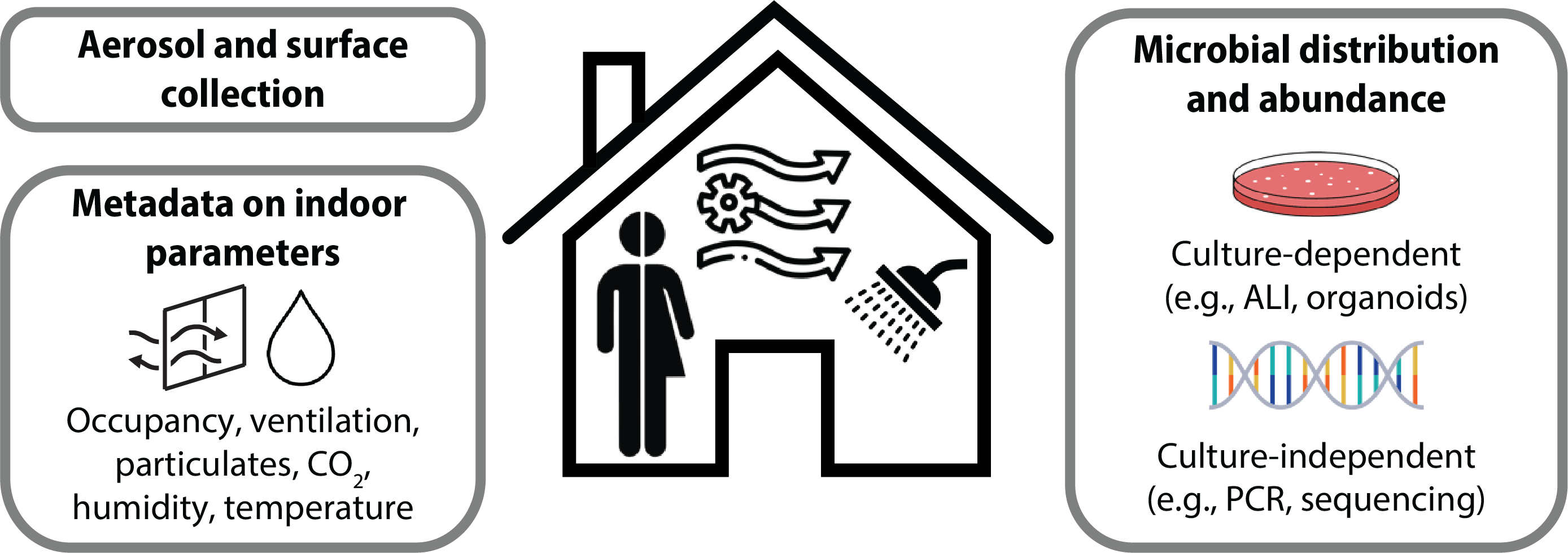
Prevalence of microbes in the built environment and informed strategies to mitigate disease
Individuals in urban settings spend over > 90% of their time indoors. Complex interactions between people and microbes in air, water, or on surfaces in these spaces can be beneficial or detrimental to human health and well-being. Characterizing the microbial composition of the built environment is therefore critical for understanding potential human health exposures and resulting acute and chronic disease risks. Yet the spatiotemporal dynamics of many microorganisms in the built environment are not known. Associations and correlations of microbes with other indoor parameters, such as CO2, occupancy, ventilation, relative humidity, and temperature, are also lacking.
We use culture dependent and independent strategies to investigate spatiotemporal dynamics of microbes, with a focus on viruses, in real-world settings. The impact of engineering controls, such as ventilation, filtration, and disinfection, is of particular interest. By understanding the prevalence of microorganisms in important indoor environments, like classrooms, homes, and clinical settings, and how they fluctuate through space and time, we can tease apart exposure risks and identify strategies that will actively shape the microbiome to improve human health.
Leveraging microbe-microbe interactions in natural and engineered systems to improve human health
There are exceptional opportunities to better understand and manipulate microbial community dynamics in engineered and natural environments to generate water or air of improved quality. We are investigating the interactions of viral and fungal populations in residential home settings, including air, water, and surfaces present in the built environment. We are particularly interested in using mycoviruses, or viruses that infect fungi, as biocontrols for limiting unwanted fungal growth and fungal byproducts (e.g., mVOCs) in these spaces. Ongoing research is centered on isolating environmental fungal strains from indoor environments for mycovirus discovery using long-read sequencing technologies, and future work will focus on establishing the impacts of virus infection on fungal virulence, growth, and metabolite production. Findings from this research will establish fungus-containing mycovirus populations that can be applied to alter the microbial community in engineered systems.

This work is supported through the NSF Precision Microbiome Engineering (PreMiEr) ERC.
